I always had a vision about computer science and technology: the advanced breakthroughs should be applied to solve real-world problems. After the breakout of COVID-19 pandemic, I believe there is no time to waste to put the computational tools to work on fighting this pandemic.
Around October in 2020, I attempted to use computational modeling to predict how COVID-19 case numbers add up with time. That was the first time I realized how simple computational tools can wonderfully work on real-life data and produce interesting results.
Figure 1. The research poster on predicting COVID-19 case number increase using a combination of different models. The research poster was accepted by IEEE-ISEC 2021.

Figure 2. The above poster won 1st place Best Poster Award in the Biology/Medicine category of the ISEC conference. (https://ewh.ieee.org/conf/stem/ISEC_2021_Best_Paper_WIP_and_Poster_Awards.pdf)
Can computational tools solve more complicated and more important problems of SARS-COV-2 virus genetic mutation? Around early 2021, news articles and research papers reported a lot of worrying discoveries. I learned how fast the virus was spreading because of mutations, which at the same time made COVID-19 escape vaccination and therefore hard to cure. I had a strong urge to find out if we can predict and thus potentially stop them. I learned interesting new science of ARIMA model and genetic mutations to worked out a model for short term mutation predictions. The manuscript of this project now appears on the preprint server bioRxiv at https://www.biorxiv.org/content/10.1101/2021.08.11.455941v4.
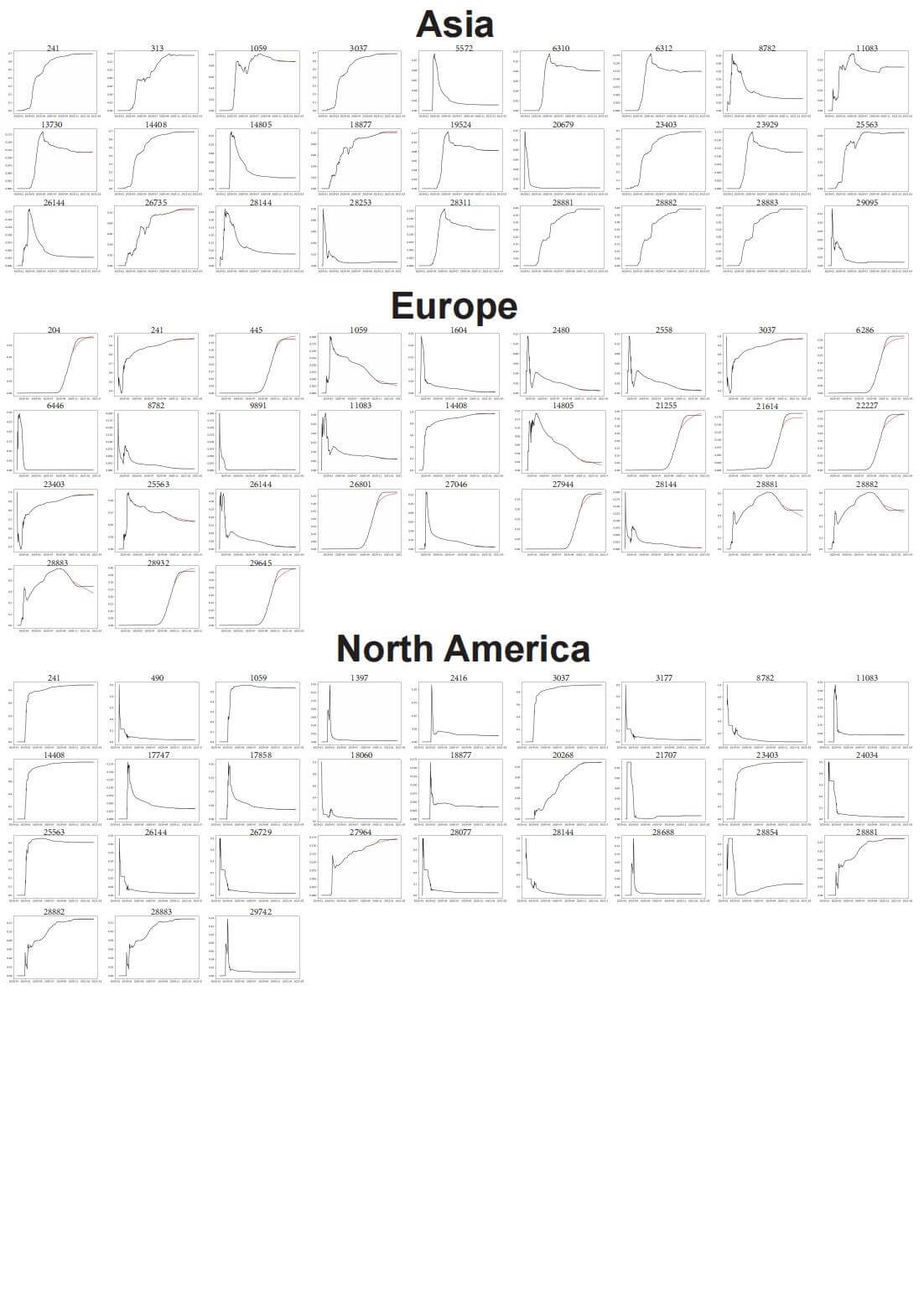
Figure 3. Prediction of genetic mutations trends for high frequency mutation sites on the SARS-COV-2 genetic sequence.
Yet the above research works are still observations and understandings of the situation. They are still far from a solution. To find a possible solution, I searched into the drug development processes for deeper understanding of the challenge. I was surprised to learn how long and costly drug developments are. Drug companies have prioritized the development of COVID-19 drugs, but their progress still is not fast enough. With knowledge from literature search and help from expert mentors, I decided to combine the latest deep learning technologies for drug repurposing predictions, which will predict whether a safe, existing drug can be used for a new disease such as COVID-19.
Figure 4. Acceptance of my research poster to the ISMB, the biggest international conference on computational biology.
Figure 5. The poster and presentation video material on ISMB virtual platform. You would need to register an ISMB account to view this material.
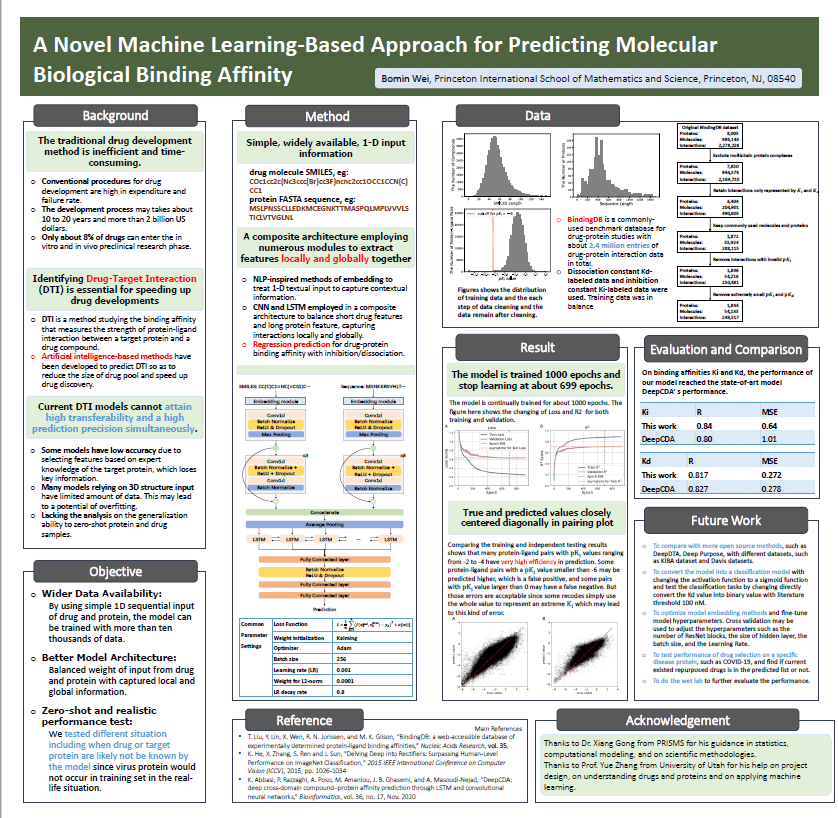
Figure 6. Research poster submitted at ISMB.
The preliminary work was accepted for a poster presentation on the international conference ISMB the biggest research conference in the field of computational biology. The work is also accepted by ECCB, which is the top European bioinformatics conference, and also ASHG, the top international bioinformatics conference. Due to COVID-19 travel concerns, I only attended ISMB in-person in July 2022 at Madison, Wisconsin.
Figure 7. Attending the ISMB in July was exciting. I talked with professionals from both research universities and industries about the drug repurposing project and learned about the research frontiers in the field.

Figure 8. A group photo at Monona Terrace, Madison, Wisconsin with the ISMB 2022 Travel Fellows
Then I spent most of the summer to optimize drug repurposing work and the most recent development of the research is accepted for a poster presentation on IDweek conference. This is a popular conference in the field of infectious disease, attended by over 8,000 experts from academia, industry, and medical sectors. The work will be presented and communicated in October, 2022, Washington D.C. Also the manuscript for this work is submitted on the preprint server bioRxiv at https://www.biorxiv.org/content/10.1101/2021.12.01.470868v2.
I envision that the developed AI drug repurposing model can be directly used by drug developers such as pharmaceutical companies and lab experts to pick the best candidates from their pool of drugs, not only for COVID-19, but also for any target protein of diseases, because the model was optimized with in mind its ability to generalize to a broad application scenario. Also, the current AI model can run predictions on a personal laptop within hours. I hope one day there will be a new drug found by my model!
